BMIQ (Beta MIxture Quantile dilation) algorithm
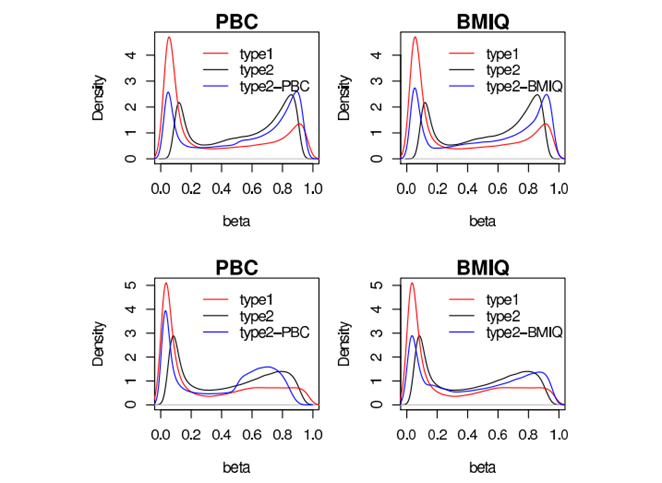
A novel model-based intra-array normalisation strategy for 450k data, called BMIQ (Beta MIxture Quantile dilation), to adjust the beta-values of type2 design probes into a statistical distribution characteristic of type1 probes. BMIQ will be useful as a preprocessing step for any study using the Illumina Infinium 450k platform.
Download and Install
Citation
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013 Jan 15;29(2):189-96.
BMIQ
R Script for BMIQ — This is the script of BMIQ
R Script for DoBMIQ — This is a script to run BMIQ on a set of methylation data.
To run BMIQ code, after placing all files (data, probe information, annotation) in one folder, you can use the following R code:
source("DoBMIQ.R")Or you can find BMIQ function in ChAMP Package, which is developed by Tiffany Morris
Installation
To install ChAMP package, start R and enter:
source("http://bioconductor.org/biocLite.R")
biocLite("ChAMP")Optional files: intro.pdf · manual.pdf