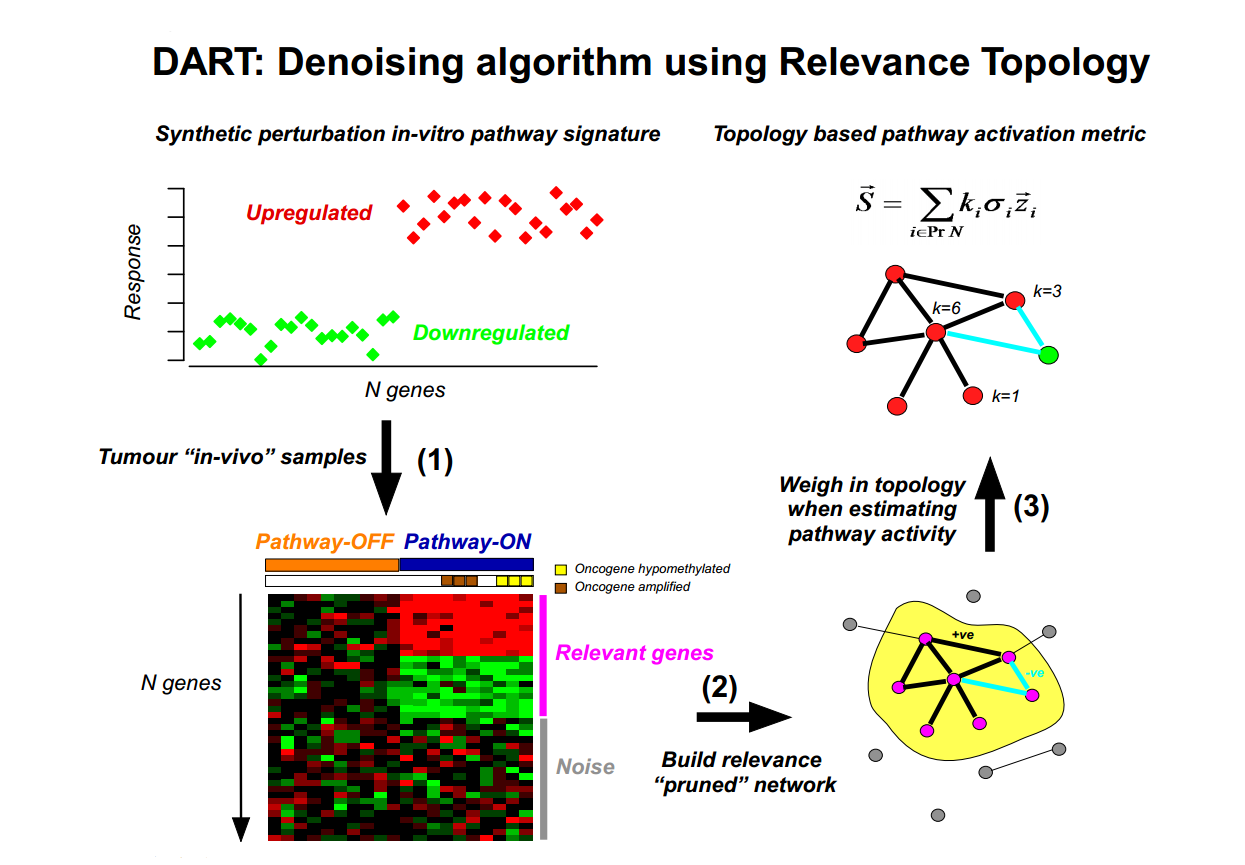
DART (Denoising Algorithm using Relevance network Topology) algorithm
Denoising Algorithm based on Relevance network Topology (DART) is an algorithm designed to evaluate the consistency of prior information molecular signatures (e.g in-vitro perturbation expression signatures) in independent molecular data (e.g gene expression data sets). If consistent, a pruning network strategy is then used to infer the activation status of the molecular signature in individual samples.
Download and Install
Citation (from within R, enter citation("DART")):
- Jiao Y, Lawler K, Patel GS, Purushotham A, Jones AF, Grigoriadis A, Tutt A, Ng T, Teschendorff AE. DART: Denoising Algorithm based on Relevance network Topology improves molecular pathway activity inference. BMC Bioinformatics 2011 Oct 19;12:403.
Maintainer: Katherine Lawler <katherine.lawler at kcl.ac.uk>
Installation
To install this package, start R and enter:
source("http://bioconductor.org/biocLite.R")
biocLite("DART")Documentation
To view documentation for the version of this package installed in your system, start R and enter:
browseVignettes("DART")PDF: DART Tutorial
PDF: Reference Manual