FEM (Functional Epigenetic Modules) algorithm
FEM can identify interactome hotspots of differential promoter methylation and differential expression, where an inverse association between promoter methylation and gene expression is assumed. A R package to identify interactome hotspots of differential promoter methylation and differential expression, where an inverse association between promoter methylation and gene expression is assumed.
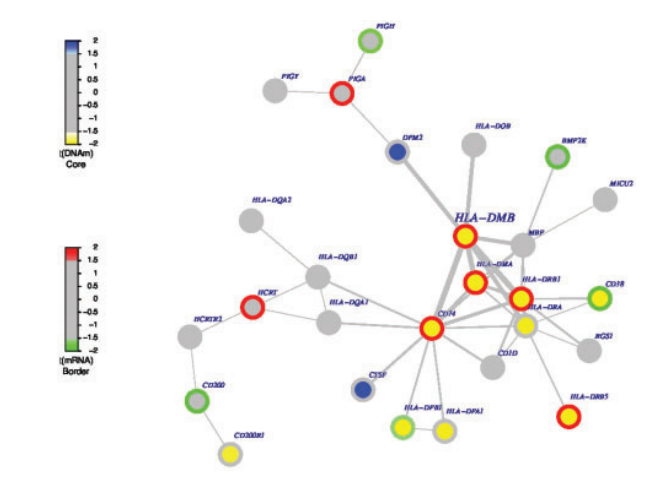
Citation
- Jiao Y, Widschwendter M, Teschendorff AE. A systems-level integrative framework for genome-wide DNA methylation and gene expression data identifies differential gene expression modules under epigenetic control. Bioinformatics 2014 Aug 15;30(16):2360-6.
- Jones A, Teschendorff AE, Li Q, Hayward JD, Kannan A, et al. (2013) Role of DNA methylation and epigenetic silencing of HAND2 in endometrial cancer development. PLoS Med 10:e1001551.
Installation
To install this package, start R and enter:
source("http://bioconductor.org/biocLite.R")
biocLite("FEM")Documentation
To view documentation for the version of this package installed in your system, start R and enter:
browseVignettes("FEM")PDF: A R package to identify interactome hotspots of differential promoter methylation and differential expression, where an inverse association between promoter methylation and gene expression is assumed.
PDF: Reference Manual