LandSCENT (Landscape Single Cell Entropy)
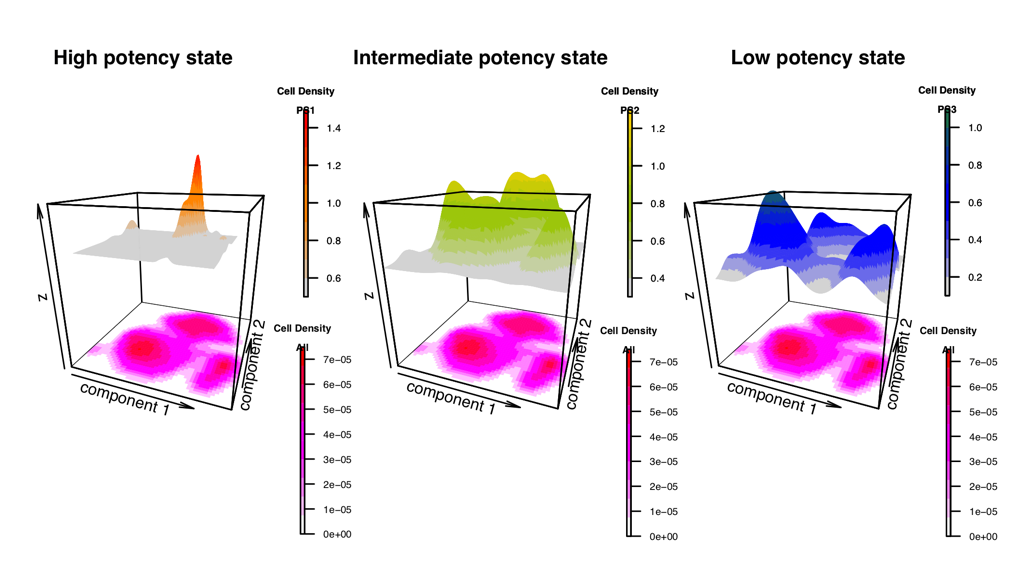
LandSCENT is an R-package for the analysis of single-cell RNA-Seq data. One main purpose of this package is to provide a means of estimating the differentiation potency of single cells without the need to assume prior biological knowledge (e.g. marker expression or timepoint). As such, it may provide a more unbiased means for assessing potency or pseudotime. LandSCENT also integrates cell density with potency distribution to dissect cell types across all potency states and generates high-quality figures to show it.
Maintainer: Weiyan Chen
Citation
- Chen W, Morabito SJ, Kessenbrock K, Enver T, Meyer KB, Teschendorff AE. Single-cell landscape in mammary epithelium reveals bipotent-like cells associated with breast cancer risk and outcome. Commun Biol 2019 Aug 9;2:306.
- Teschendorff AE, Enver T. Single-cell entropy for accurate estimation of differentiation potency from a cell's transcriptome. Nat Commun 2017 Jun 1;8:15599.
- Teschendorff AE, Sollich P, Kuehn R. Signalling entropy: A novel network-theoretical framework for systems analysis and interpretation of functional omic data. Methods 2014 Jun 1;67(3):282-93.
- Chen W, Teschendorff AE. Estimating Differentiation Potency of Single Cells Using Single-Cell Entropy (SCENT). Methods Mol Biol 2019;1935:125-139.
- Shi J, Teschendorff AE, Chen W, Chen L, Li T. Quantifying Waddington's epigenetic landscape: a comparison of single-cell potency measures. Brief Bioinform 2020 Jan 17;21(1):248-261.
Installation
To install this package, start R and enter:
if (!requireNamespace("devtools", quietly = TRUE))
install.packages("devtools")
devtools::install_github("ChenWeiyan/LandSCENT")Documentation
HTML: LandSCENT Vignette