Signalling Entropy
This software implements a sample-level signalling entropy framework. Gene expression data should be positively valued (e.g. Affy/Illumina intensities or RNA-Seq count data). The mass action principle is used to construct the stochastic matrix over a protein interaction network.
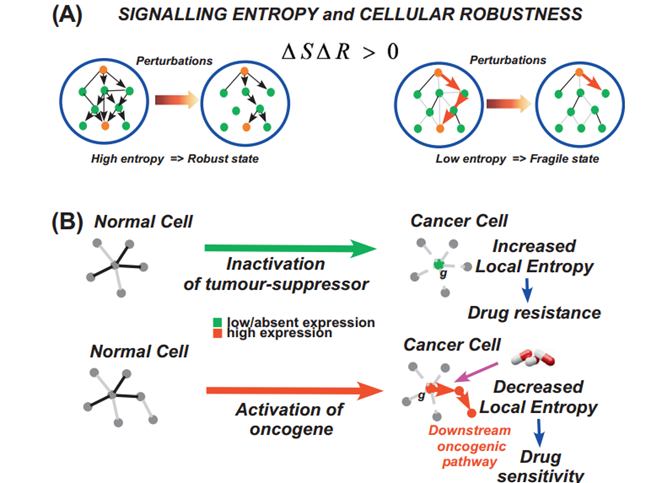
Citation
- Teschendorff AE, Sollich P, Kuehn R. Signalling entropy: A novel network-theoretical framework for systems analysis and interpretation of functional omic data. Methods 2014 Jun 1;67(3):282-93.
Description of files
1) dataSCM2.Rd: R data object containing gene expression matrix avdataSCM2.m (≈17,000 genes × 191 samples) from the Stem Cell Matrix 2 compendium. Cell type is in phenoSCM2.lv[[1]] (hESC=1, iPSC=2, differentiated=3). Row names are Entrez Gene IDs.
2) hprdAsigH-13Jun12.Rd: R data object containing a PPI network adjacency matrix hprdAsigH.m. Row names are Entrez Gene IDs.
3) sr.Rd: Auxiliary R object needed to run the vignette.
4) DoIntegPIN.R: R function to integrate a PPI network with a gene expression matrix and extract the maximally connected subnetwork.
5) CompSR.R: R function to compute the entropy rate (plus auxiliary functions).
6) CompSRana.R: R function to compute the entropy rate analytically assuming detailed balance (faster computation).
7) PDF: Vignette / manual.